NL Journal of Agriculture and Biotechnology
(ISSN: 3048-9679)
Utilizing Biotechnology for Plant Disease Management: A Comprehensive Review
Author(s) : Manjot Kaur, Muskan Bhullar*, Ramneek Kaur. DOI : 10.71168/NAB.02.06.134
Abstract
Threat to agriculture economy from pests and diseases are increasing constantly. Global food crops are lost from 20% to 40% due to pests and diseases, with damage reaching 14.1% by plant diseases, accounting for $ 220 billion in annual agricultural trade losses. Traditional methods like chemical control have a great detrimental effect on environment. Thus, an alternative concept of Molecular detection techniques and biotechnology sets diagnostic tools, techniques can generate accurate results rapidly enough to be useful for disease management decisions. Biotechnology is the genetic manipulation, and multiplication of any living organism through novel techniques and technologies such as tissue culture and genetic engineering resulting in the production of improved or new organism and products that can be used in variety of ways. The various applications of biotechnology in plant pathology includes, Genetic Resistance, Marker Assisted Breeding, Genetic Engineering, RNA Interference, Gene Pyramiding and tagging of resistant genes. Keywords: Agriculture, Biotechnology, Disease Management, Genetic Resistance, Molecular Detection, RNA Interference.
Introduction
The plants are under constant and increasing threat from pests and diseases. According to the Secretariat of the International Plant Protection Convention (IPPC) under the FAO, if no actions and measures are taken in a timely manner against pests and diseases, the situation can lead to tragic and difficult consequences. Today, global food crops are lost from 20% to 40% due to pests and diseases, with damage reaching 14.1% by plant diseases, accounting for $ 220 billion in annual agricultural trade losses [2]. Accurate identification and early detection of pathogens is the cornerstone of disease management in many crops. Traditional plant breeding methods have been used to develop cultivars resistant to various diseases [1]. Traditional methods like chemical control is often non-specific in its effects, killing beneficial organisms as well as pathogens, and it may have undesirable health, safety, and environmental risks. Because of these factors, nowadays, control of plant disease is a subject of great interest for biotechnologists [7,24]. Thus, an alternative concept of Molecular detection techniques and biotechnology sets diagnostic tools, techniques can generate accurate results rapidly enough to be useful for disease management decisions. The application of these technologies in plant pathology has greatly improved our ability to detect plant pathogens and is increasing our understanding of their ecology and epidemiology [15].
Biotechnology involves the use of living organisms in industrial processes particularly in agriculture, food processing and medicine [25]. According to Agrios [2] biotechnology is the genetic manipulation, and multiplication of any living organism through novel techniques and technologies such as tissue culture and genetic engineering resulting in the production of improved or new organism and products that can be used in variety of ways. Biotechnology is derived from the fusion of two words “Biology and Technology” [6]. According to U.S. National Foundation Biotechnology is defined as the controlled use of biological agents, such as, microorganisms, or cellular organisms for beneficial use. Plant biotechnology uses non-conventional approaches based primarily on cell and tissue culture for either improving the genetic makeup, phenotypic performance or multiplication rates of economic plants, or at exploiting plant cell and cell constituents for generating useful products or/and services such as diseases resistance [10].
Applications of biotechnology in plant pathology
A. Genetic resistance
Plants have evolved a variety of activities and passive mechanisms to defend themselves against pathogens, and disease resistance genes have been incorporated into crop plants to protect them against diseases caused by the pathogens. Development of resistant plants depends on the tissue culture technology in which vegetative parts are used to produce plants that are genetically identical. Following are the methods through which disease resistance is developed in plants:
a. Disease resistant mutants from plant cell cultures
The cells of and the plant regenerated from cell culture show heritable variation for both quantitative and qualita- tive traits; such variation is known as somaclonal variations. These variants can also be obtained for disease resist- ance. Many pathogenic bacteria and fungi produce either specific or non-specific toxins. Plant cell culture has been exposed the sublethal concentration of the specific toxins and resistant clones isolated. Plant regenerated from resistance clones are resistant to the disease producing pathogen. For example, maize lines having Texas male sterility are susceptible to Helminthosporium maydis which produce toxin that binds to mitochondria. Maize cells resistant to this toxin have been selected and plant regenerated from them were resistant to southern leaf blight. This approach is successful only in the case of pathogens producing specific toxins or toxins that are involved in disease development. Further, many of the pathogens do not produce a toxin or produce non-specific toxins. In cases of such pathogen plantlets may be regenerated from unselected cell culture, and the R1 generation of these plants may be screened for resistance to concerned pathogen. This approach was successful in the isolation of bacterial wilt resistance tomato line and of Fiji disease resistant sugarcane line (c.v. Ono) [21].
b. Resistant double haploids from haploid plants
Haploid plants can be developed by inducing immature pollen cells or sometimes megaspores of many plants. Haploids are plants (sporophytes) that contain a gametic chromosome number (n). They can originate sponta- neously in nature or because of various induction techniques. Spontaneous development of haploid plants has been known since 1922, when Blakeslee first described this phenomenon in Datura stramonium [3]. Following regeneration, haploid plants obtained from either anther or ovule culture may grow normally under in vitro con- ditions Most highly resistant haploids can be sorted by vegetative propagation and proper screening. Selected plant treated for diploidization of nuclei. Various methods have been applied over several decades and are still in development. The most frequently used application is treatment with anti-microtubule drugs, such as colchicine (originally extracted from autumn crocus Colchicum autumnale), which inhibits microtubule polymerization by binding to tubulin. Other options are oryzalin, amiprophosmethyl (APM), trifluralin and pronamide, all of which are used as herbicides and are effective in micromolar concentrations [21].
Popular example of double haploid resistant plant is Barley possessing resistance to stem rust, leaf rust and powdery mildew through DH [23].
c. Increase in disease resistance by protoplast fusion
Production of hybrid plants through the fusion of protoplasts of two different plant species/varieties is called somatic hybridization, and such hybrids are known as somatic hybrids. The technique of somatic hybridization involves the following four steps. 1. Isolation of protoplasts, 2. Fusion of protoplasts 3. Selection of hybrid cells, and 4. Proliferation of the hybrid cells and regeneration of hybrid plants from them.
Protoplast fusion is particularly useful between protoplast of different, highly resistant haploid lines in the same variety or species, and it can result in diploid plants that combine the resistant genes of two highly resistant haploid lines [6]. By this method, factors that contribute to crossing barriers between species can be avoided and viable hybrids (Cybrids) have been recovered even between distantly related species. The application of somatic hybridization is restricted mainly to solanaceous and crucifer crops. Even within these two families, exploitation of this technology is relatively limited. Two tobacco varieties Delgold and Dr. Chang, developed from protoplast fusion between N. tabacum and N. rustica have been released in Canada. Examples of disease resistant plants, produced from protoplast fusion are shown in Table 1 below.
d. Chemically induced fusion
Isolated protoplasts are sticky, tend to aggregate in suspension and show fusion spontaneously during incuba- tion. Chemicals tend to increase the fusion frequency. Fusion can occur in the presence of high CA2+ and high pH [9-10] but a commonly used chemical (Fusogen) is polyethylene glycol (PEG). Due to the addition of PEG, there is adhesion of protoplast to their neighbors which can be assessed by microscope. Subsequent dilution of stabilized PEG, either stepwise or at once results in fusion and mixing of the cytoplasm. PEG causes slight dehydration of the protoplasts and crinkling of the membrane. The level of fusion is usually 1-10% as chemical fusion agents are toxic and therefore damaging to the cell [11].
Table 1: Disease resistant plants produced from protoplast fusion
| Species used for fusion | Disease | Reference |
| Lactusa sativa | Downy mildew (Bremia lacticae) | Maloy, 2005 [14] |
| Brassica oleraceae and Raphanus sativus | Club root (Plasmodiophora brassical) | Maloy, 2005 [14] |
| Brassica napus and Brassica nigra | Black leg (Phoma lingum) club root | Maloy, 2005 [14] |
| Solanum brevidens and Solanum tuberosum | Bacterial soft rot (Erwinia spp) | Maloy, 2005 [14] |
B. Marker-Assisted breeding
Marker-Assisted breeding or Marker-Assisted selection is indirect selection of gene/QTL based on molecular marker closely linked to gene/QTL (foreground selection) Table 2. Molecular marker distributed throughout the genome can be used to aid the recovery of genomic region, except for the target region of one of the parents (background selection). MAB enhances plant breeding by efficient incorporating disease resistance traits using molecular marker (Marker-Assisted Backcrossing). Process of Marker-Assisted breeding includes Marker Iden- tification (Identify markers associated with disease resistance genes), Genotyping (Assess genetic makeup of plants using markers), Linkage Mapping (Map markers relative to disease resistance genes), MAS (Select plants with marker-linked disease resistance traits).
A four-stage sampling strategy, as outlined below, has been proposed for selection objective:
- Selection of individual carrying target allele (gene under transfer, foreground selection).
- Selection of individuals, from among the above, that are homozygous for the recurrent parent genotype at loci flanking the target locus.
- Selection of individual (from ii) homozygous for recurrent parent genotype on the remaining loci of the chro- mosome having the target gene.
- Selection of at least on individual (from iii) that is homozygous for the recurrent parent genotype at the max- imum number of loci.
For Example: Fusarium head blight (FHB) in wheat. A major QRLFhb1for resistance to FHB (causal agent Fusarium graminearum) was mapped on wheat chromosome 3B in a population derived from resistant parent cultivar ‘Sumai 3 (Singh, 2015).
Table 2: Depicts MAS for gene transfer through backcross programme [5].
|
Crop |
Character |
Gene/QTL |
MAS | |
| Foreground | Background | |||
|
Barley |
Barley Yellow Dwarf Virus resistance
Leaf rust resistance |
Yd2
Rphq6 |
STS
AFLP |
None
AFLP |
|
Maize |
Corn borer resistance |
QTLS on chromosome 7, 9 and 10 |
RFLP |
RFLP |
|
Rice |
Bacterial leaf blight resistance
Blast resistance |
Xa21, xa5, xa13
Pi1 SSR |
STS, SSR or CAPS
SSR |
RFPL, AFLP or none |
|
Wheat |
Powdery mildew resistance
Stripe rust resistance |
22 Pm genes
Yr 10, Yr 15 |
Phenotyping
SSR |
SSR
SSR |
C. Genetic engineering
The most potent biotechnological approach for creating the desired genetic variation is the transfer of specifically constructed gene assemblies through various transformation. The plant obtained from genetic engineering con- tain a gene or genes usually from an unrelated organism; such gene is called transgene, and the plant containing them are known as transgenic plants. The production of transgenic plants is achieved by recombinant DNA tech- nology that enables the transfer of transgenes into plant cells and integrate them into plant genomes. For disease resistance genes confers disease resistance are isolated, cloned and transferred into the crop in question.
Genetic engineering for disease resistance in plants is done in two ways:
a. Genetic engineering and artificial cell death
In this method hypersensitivity response is mimicked. The interaction of pathogens with plants leads to a disrup- tion in cellular homeostasis, often leading to cell death, in both compatible and incompatible relationships. The mechanistic basis of this cellular disruption and consequent death is complex and poorly characterized, but it is established that host responses to pathogens are dependent on gene expression, involve signal transduction, and require energy. It is brought about by endogenous gene action particularly in response to some specific stimulus. Artificial programmed cell death covers all the races of the pathogen as well as more than one pathogen. There are two schemes followed for artificially programmed cell death.
- Two component system: Two precisely matched genes (avr gene and R gene) are expressed in same avr gene is driven by a promoter induced non-specific elicitor during pathogen attack. R gene is used correspond- ing to the avr gene. The expression of avr gene must be expressed immediately following pathogen attack but only in infected cells. Elicitor produced by avr is recognized by R gene leading to plant cell death.
- Single component system: Based on expression of polypeptide in response to pathogen infection. Trans- genes used here are those that encodes toxins ribonucleases, or other enzymes whose product are toxic to plant cells leading to Example of PCD Barnase gene from B. amyloliquefaciens was transferred into po- tato for the control of Phytophthora infestans.
b. Pathogen derived resistance
Based on gene taken from genome of virus pathogen and engineered into the host. First introduced in tobacco by Roger Beachy and coworkers. It is well established that plants inoculated with a mild strain of a virus become ‘resistant’ to a subsequent infection by a ‘virulent’ strain of the same virus; this phenomenon is known as virus cross-protection. In most cases of cross-protection, the development of symptoms following the infection by a virulent strain of the virus is markedly delayed most likely due to a suppression of its replication. The successful approaches for transgenic virus resistance are transfer of the coat protein gene of a virus into the genome of its host, where it is constitutively expressed. For example, the coat protein gene of TMV has been transferred into the tobacco genome; the transgenic plants showed expression of this gene, and very low and delayed symptom development on inoculation with TMV. This approach has been used to develop ‘virus protected’ varieties of some crops [21].
Another approach is to transfer cDNA (complementary DNA) of satellite RNA (sat-RNA) of those RNA viruses that contain an appropriate sat-RNA. Satellite RNA replicates with the viral genome, gets packed into the virus parti- cle along with the latter, and reduces, increases or does not affect the severity of symptoms due to those strains which carry it. Therefore, DNA complementary to such sat-RNAs that reduce the severity of symptoms produced by RNA viruses have been transferred into the genomes of their hosts in the hope of conferring upon them re- sistance to infection by that virus. In all such cases, the transgenic plants showed a much lower level of symptom development than the respective controls when inoculated with sat-RNA free highly virulent strains. In addition to these, some other strategies for conferring virus resistance into plants are being pursued, e.g., transfer of genes encoding antibodies against viruses (success achieved in potato), antisense RNA approach, ribozyme-mediated protection, etc. But so far, coat protein gene is the only strategy for commercial production of disease resistance varieties [20]. Some types with example of pathogen derived resistance; Coat protein-mediated resistance (for tobacco mosaic virus), Replicase-mediated resistance (cucumber mosaic virus), Nucleic Acid-mediated resist- ance (turnip yellow mosaic virus), Movement protein mediated resistance (Tomato mottle virus ToMoV).
D. RNA Interference (RNAi)
RNA interference (RNAi) or Post-Transcriptional Gene Silencing (PTGS) is a conserved biological response to double-stranded RNA that mediates resistance to both endogenous parasitic and exogenous pathogenic nucleic acids, and regulates the expression of protein-coding genes. RNAi-related pathways in plants give rise to different types of sRNAs [22].
Mechanism of RNAi
Long dsRNA (which can come from hairpin, complementary RNAs, and RNA-dependent RNA polymerases) is cleaved by an endo-ribonuclease called Dicer. Dicer cuts the long dsRNA to form short interfering RNA or siRNA and in miRNA Drosha help in cleavage; this is what enables the molecules to form the RNA-Induced Silencing Com- plex (RISC). Once siRNA/miRNA enters the cell it gets incorporated into other proteins to form the RISC. Once it is part of the RISC complex, it is unwound to form single stranded siRNA/miRNA. The single stranded siRNA/miRNA which is part of the RISC complex now can scan and find a complementary mRNA. Once the single stranded siRNA/ miRNA (part of the RISC complex) binds to its target mRNA, it induces mRNA cleavage. The mRNA is now cut and recognized as abnormal by the cell. This causes degradation of the mRNA and in turn no translation of the mRNA into amino acids and then proteins. Thus, silencing the gene that encodes that mRNA [17].
Applications in Plant Disease Management
Viral Diseases: RNAi has been successfully applied to combat viral infections in plants. By targeting specific viral genes, it can hinder viral replication and spread within plant tissues.
Fungal and Nematode Pathogens: RNAi can also be employed against fungal and nematode pathogens by si- lencing essential genes in these organisms, disrupting their ability to infect plants. The first RNAi translational applications were for plant virus disease control. Transgenic squash, papayas, and potatoes were first commer- cialized in the United States in mid 1990s. At about this same time, peppers and tomatoes with transgenic re- sistance to Cucumber mosaic virus (CMV) were deregulated in China. The next approvals for commercial use of RNAi-based plant virus control were not until almost 10 years later for plums in the United States against Plum pox virus (PPV), and beans in Brazil against Bean golden mosaic virus (BGMV).
E. Gene Pyramiding
Bringing together two or more genes governing a single trait, especially resistance to disease, is known as gene pyramiding. In case of resistance breeding, gene pyramiding involves bringing two or more distinct genes con- ferring resistance to a single pathogen in the same line/variety. Gene pyramiding using conventional approaches is quite cumbersome and involved, as it requires extensive progeny tests and repeated disease tests. The use of molecular markers greatly facilitates gene pyramiding as it minimizes the need for disease tests and progeny tests. Figure 1 show gene pyramiding: For instance, gene Xa21 for resistance to bacterial leaf blight (BLB) of rice generates resistance to most of the BLB races. STS markers have been used for MAS to pyramid resistance genes Xa4, xa5, xa13 and Xa21 in all possible combinations; the lines derived from the programme show a wider spectrum or higher level of resistance to BLB. Two rice varieties, ‘Angke’ and ‘Conde’ developed by MAS for BLB resistance have been released in Indonesia. Similarly, MAS has been used to pyramid genes Pil, Piz5 and Pita for resistance to rice blast (Magnaporthe grisea). In India, Pusa Basmati 1 has been improved by transferring bac- terial leaf blight resistance genes xa13 and Xa21 using MAS in a backcross-pedigree programme. The improved version has been released as Improved Pusa Basmati 1, which is the first example of a variety developed through MAS in this country.
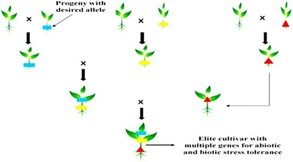 Figure 1: Gene pyramiding flowchart
Figure 1: Gene pyramiding flowchart
F. Molecular tools for disease diagnosis
Accurate identification and early detection of pathogens is the cornerstone of disease management in crops. Technologies for the molecular detection of plant have already undergone two major breakthroughs over the past 30 years [12]. The first was the advent of antibody-based detection, in particular monoclonal antibodies and enzyme-linked immunosorbent assay [4]. Then came DNA-based technologies, such as the polymerase chain re- action (PCR), Nucleic acid hybridization, molecular markers and microarrays; these tools for pathogen detection are described further:
a. Polymerase chain reaction (PCR)
The polymerase chain reaction (PCR) provides a simple, ingenious method to exponentially amplify specific DNA sequences by in vitro DNA synthesis. Three essential steps to PCR include (a) melting of the target; degradation (b) annealing of two oligonucleotide primers to the denatured DNA strands, and (c) primer extension by a ther- mostable DNA polymerase shown in Figure 2. Newly synthesized DNA strands serve as targets for subsequent DNA synthesis as the three steps are repeated up to 50 times. The specificity of the method derives from the synthetic oligonucleotide primers, which base-pair to and define each end of the target sequence to be amplified. The use of PCR grew rapidly in plant pathology, as in other disciplines. PCR offers several advantages compared to more traditional methods of diagnosis: organisms need not be cultured prior to their detection by PCR. The technique possesses exquisite sensitivity, with the theoretical potential to detect a single target molecule in a complex mixture without using radioactive probes; and it is rapid and versatile. Like serology, both narrow and broad selectivity is possible and, depending on the choice of primers, the method facilitates the detection of a single pathogen or many members of a group of related pathogens [8].
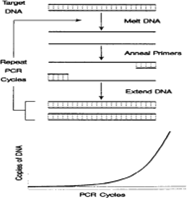 Figure 2: Depicts stages of PCR and the resultant amplification of DNA copies of the target region.
Figure 2: Depicts stages of PCR and the resultant amplification of DNA copies of the target region.
b. Molecular Hybridization
The basic process of binding a single stand of nucleic acid (DNA or RNA) to its complementary strand is called Nucleic Acid Hybridization. Procedure includes four steps; (i) denaturation, separation of DNA strands by heat or chemicals and preparing single-stranded DNA or RNA for hybridization; (ii) Hybridization, Combining target DNA/RNA with a complementary probe. Facilitating the formation of stable duplexes between pathogen nucleic acid and specific probes; (iii) Washing, Removing unbound probes. Enhancing specificity by eliminating non-spe- cific binding. (iv) Detection, Identifying hybridized DNA/RNA. Visualizing or quantifying the presence of patho- gens in the sample. Molecular hybridisation-based assays were first utilised in plant pathology to detect Potato spindle tuber viroid [16] and adapted to virus detection [9]. However, certain problems associated with the use of radioactive probes, relatively low sensitivity and complexity of these techniques and the development of amplifi- cation-based assays have minimized new improvements and applications. The most common molecular hybridi- zation format for the detection of viruses is non-isotopic dot-blot hybridization using digoxigenin-labelled probes. This technique has been employed for Apple mosaic virus (ApMV), Prunus necrotic ringspot virus (PNRSV), Prune dwarf virus (PDV), PPV, and Apple chlorotic leaf spot virus (ACLSV). Molecular hybridisation can also be applied to the specific detection of amplicons generated after amplification techniques based on PCR, thereby increasing their sensitivity and specificity levels [26] and reducing time when a flow-through system is used.
c. Molecular markers
Molecular markers consist of specific molecules, which show easily detectable differences among different strain of species or among different species. These markers may be based or proteins (isozymes) or DNA. DNA-based markers comprise a variety of markers based on difference in DNA sequences and are extremely versatile. In cur- rent usage molecular markers signifies DNA markers. There are several DNA-based marker systems, which are based either on analysis of restriction enzyme digests of genomic DNA or of amplification products generated by PCR (polymerase chain reaction); some markers combine both these features. The different molecular markers are classified as DNA hybridization-based, PCR-based and sequence-based makers based on the method of their detection and, more particularly, analysis. Restriction fragment length polymorphism (RFLP) and diversity array technology (DART) are examples of hybridization-based markers. Common examples of PCR-based markers are random amplified polymorphic DNAs (RAPDs), sequence characterized amplified regions (SCARs), inter-sim- ple sequence repeats (ISSRs), simple sequence repeats (SSRs), cleaved amplified polymorphic sequences and start codon-targeted (SCoT) markers. The marker systems amplified fragment length polymorphism (AFLP) and cleaved amplified polymorphic sequences (CAPSs) share some features of both RFLP and RAPD marker systems. Single nucleotide polymorphism (SNP) is the only sequence- based marker system. Molecular markers have var- ious applications in detection of plant pathogens. In the late 1990s, RAPD markers were utilized to study the genetic diversity of Phytophthora infestans. SCAR markers have been employed in the identification of Fusarium oxysporum f. sp. cubense (tropical race 4), the causative agent of Panama disease in bananas. Various strains of Xanthomonas axonopodis pv. citri, the bacterium responsible for citrus canker analysed by RFLP patterns, re- searchers could distinguish between different pathogenic strains. LAMP was used for the detection of the causal agent of downy mildew in grapes, Plasmopara viticola.
d. Microarray technology
Microarrays are generally composed of thousands of specific probes spotted onto a solid surface (usually nylon or glass). Each probe is complementary to a specific DNA sequence (genes, ribosomal DNA) and hybridization with the labelled complementary sequence provides a signal that can be detected and analysed. Since the development of microarray technology for gene expression studies [19], new approaches are extending their application to the detection of pathogens. Although there is great potential for microarray technology in the diagnosis of plant diseases, the practical devel- opment of this application is still in progress. the microarray technology focuses its use on multiplex format of similar or very different pathogens, taking advantage of the number of probes that can be employed in one chip. With the availability of genomic sequences of pathogens and the rapid development of microarray technology, as well as a renewed emphasis on detection and characterization of quarantine pathogens. The potential of mi- croarray technology in the detection and diagnosis of plant diseases is very high, due to the multiplex capabilities of the system. Moreover, it can be coupled with other systems, i.e. to perform nucleic-acid extraction on the chip achieve PCR reactions and their detection on the same device achieve PCR reactions and their detection on the same device providing the possibility of automation that can be of great importance and utility. This possibility, with the coupling with previous steps of the analyses (extraction, PCR, detection) promises a wider use in future protocols. Furthermore, new developments, like the labelling of total bacterial RNA, the direct detection of DNA or RNA without previous PCR amplification, may make this technique simpler [13].
G. Tagging of resistance genes
Tagging refers to the mapping of disease resistance genes close to known markers. Such tags can be used for indirect selection of such genes in breeding programmes. Plants can recognize and resist invading pathogens by signaling the induction of rapid defense responses. Often these responses are mediated by single dominant resistance genes (R genes). The products of R genes have been postulated to recognize the pathogen and trigger rapid host defense responses (Kumar and Whitham et al., 1995).
Table 2: Depicts MAS for gene transfer through backcross programme [5].
| Crop | Name if trait / gene tagged |
|
Wheat |
Leaf rust resistance genes Lr9, Lr10, Lr21, 3Lr24, Lr37, Lr34, Lr46
Stem rust resistance genes Sr21, Sr33 Powdery mildew resistance genes Pm1, Pm2, Pm8, Pm12 |
| Rice | Blast resistance genes Pi2(t), Pi4(t)
Bacterial blight resistance genes Xa1, Xa2, Xa3, Xa4 and Xa21 |
Example, two RAPD markers linked to resistance (assayed as total girdling of stalk by black leision) to stalk rot (Sclerotinia sclerotiorum) in cauliflower were identified using a mapping population of 200 F₂ (Olympus × PSB) plants with Olympus being the resistant parent. The analysis of genetics of stalk rot resistance was based on the parents and F₂ generation. Stalk rot resistance in cauliflower appeared to be governed by many genes (polygenic character). A total of 222 random decamer primers were subsequently used to survey the parental polymor- phism regarding DNA amplification by Polymerase Chain Reaction. The primers which showed polymorphism in parental lines were used for bulked segregant analysis. Those that amplified consistently and differentially in the resistant and susceptible bulks were used for single plant analysis of 200 F₂ plants. RAPD markers D-3₄₅₀ (5 GGACCCAACC 3) and C-20₃₅₀ (5 ACTTCGCCAC3) flanking the stalk rot resistance gene with 2.7 cM, and 42 cM, respectively, were identified. These two markers are close enough to the stalk rot resistance gene to allow a de- pendable marker-assisted selection for stalk rot resistance [18].
Conclusion
Biotechnology is the genetic manipulation, and multiplication of any living organism through novel techniques and technologies such as tissue culture and genetic engineering resulting in the production of improved or new organism and products that can be used in variety of ways. The various applications of biotechnology in plant pathology includes, Genetic Resistance, Marker Assisted Breeding, Genetic Engineering, RNA Interference, Gene Pyramiding and tagging of resistant genes.
References
2. Agrios G N (2005) Plant Pathology. Academic Press, ISBN 978-0-12-04456-3.
This article licensed under the Creative Commons Attribution 4.0 International License CC-BY 4.0., which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are properly credited.
